Session 1
Created Oct 2022, Last updated: 12 December, 2024
What is this page for?
This page has code for session 1 of BPI stats workshop. This session mainly focusses on R programming and is for beginners. We might not finish all of this in the session, but you will have the material for use later.
These exercises will only work if you have completed the pre-session items described here correctly. After following instructions on this page, go to the Session 2 page.
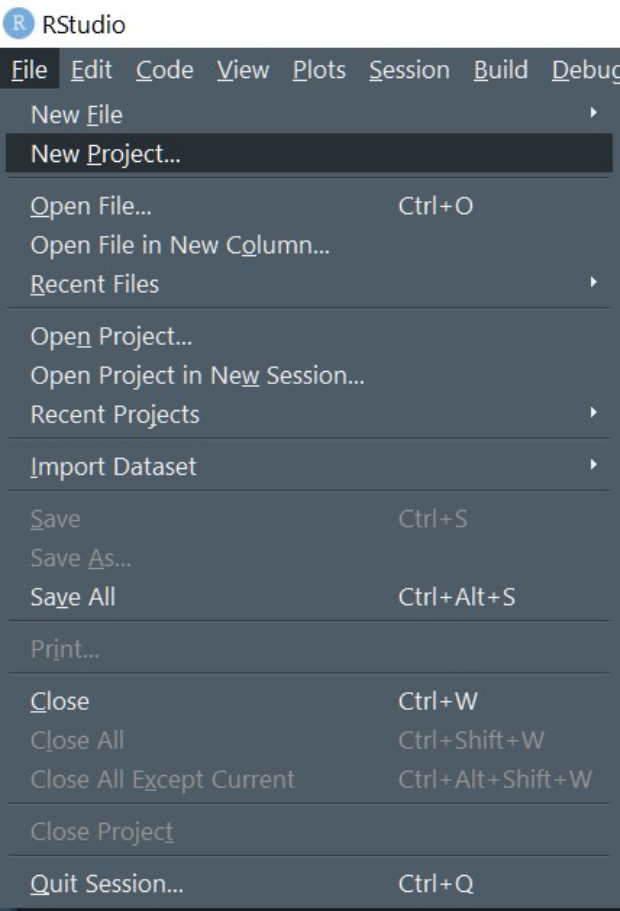
Create a new Project
Keep your work organised in RStudio by working within “Projects”. Start by creating a new project in a directory you want to save your work in.
Create a new project by using File > New Project menu in RStudio.
The new project dialogue box looks like this on Windows. Create a New Directory (e.g., Rstats_session1).
RMarkdown file
This is the best way to save analyses, including code and its output (e.g., graphs), along with formatted text. Outputs can be saved as Word, PDF or HTML files (HTML recommended).

A website like this one, which contains rich text, code and output, can be created entirely in RMarkdown format.
In RMarkdown files, writing *italics* converts text into italics, **bold** into bold and so on. This allows ‘rich text’ notes (e.g., to describe experiments and results).
R code is enclosed within ```{r}```
(three back ticks, curly bracket, small r followed by lines of code that ends with a new line of curly bracket and three back ticks). These are called code chunks.
Here are more examples from the RMarkdown cheatsheet.

Create your own RMarkdown result of this session
There will be an error if you have not installed all the packages required to knit an R Markdown file; first install the packages, then follow the code below.
To do this, create a new RMarkdown file by going to File -> New File -> R Markdown. This should open a new dialogue box in which you can name the new file as “Session1” and click OK.
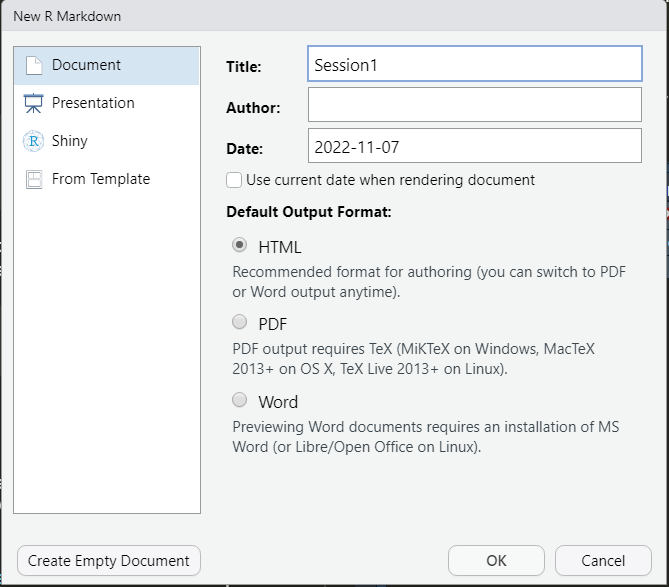
The new file is a generic template where the header with file details (title, output and date) is between three dashed lines --- at the top. This is called the YAML metadata or YAML header.
Text below this is code to be executed or just descriptive text. When the code and text is ready, use the Knit button on top of the pane to ‘knit’ the file into an HTML document (save it with a name such as “Session1.html”)

Create your RMarkdown file and write rest of today’s code within R chunks. This way you will have a record of the code and its output from this session.
The basic operators
Let’s look at the common operators in R.
- assign operator
<-(you can also use=). PressingAltand-together is the short-cut for this operator. You will use<-to assign values and create objects in R. The<-can also be used to generate and save new columns in a data table (see step 6 below).
my_first_object <- "A"
my_second_object = "B"- common arithmetic operators:
*,/,+,-,^, and brackets(). BODMAS rule applies in order of processing.
5*2^2 [1] 20#same as
5*(2^2)[1] 202/10[1] 0.23*20-10[1] 503*(20-10)[1] 302^2*(5-3)[1] 82^2*5-3[1] 17- hash
#to comment out a line or add comments after lines of code. For example, typing5*6on the console will give you the result (30), but# 5*6will not work because R thinks that’s a comment.
#multiplication 5*2
5 * 20 #5 multiplied by 20[1] 100- square brackets
[]have a special meaning. They are used for sub-setting data frames (i.e., only picking rows in the table that match a criterion).
#pick the cell in row 2 and column 1
cars[2, 1][1] 4#pick rows 2, 3, 4 and all columns 2
cars[c(2,3,4), 2][1] 10 4 22#to pick all columns, leave blank after comma
cars[c(2,3,4), ] speed dist
2 4 10
3 7 4
4 7 22#pick rows 1-10 in all columns
cars[1:10,] speed dist
1 4 2
2 4 10
3 7 4
4 7 22
5 8 16
6 9 10
7 10 18
8 10 26
9 10 34
10 11 17- equal
=operator is used to assign anarugmentof afunction.
tail(cars, n = 10) speed dist
41 20 52
42 20 56
43 20 64
44 22 66
45 23 54
46 24 70
47 24 92
48 24 93
49 24 120
50 25 85- dollar
$operator is used to look up columns in a table. A double bracket[[ ]]can also be used here.
cars$speed [1] 4 4 7 7 8 9 10 10 10 11 11 12 12 12 12 13 13 13 13 14 14 14 14 15 15
[26] 15 16 16 17 17 17 18 18 18 18 19 19 19 20 20 20 20 20 22 23 24 24 24 24 25cars[["speed"]] [1] 4 4 7 7 8 9 10 10 10 11 11 12 12 12 12 13 13 13 13 14 14 14 14 15 15
[26] 15 16 16 17 17 17 18 18 18 18 19 19 19 20 20 20 20 20 22 23 24 24 24 24 25Operators can be combined.
cars$speed * 20 [1] 80 80 140 140 160 180 200 200 200 220 220 240 240 240 240 260 260 260 260
[20] 280 280 280 280 300 300 300 320 320 340 340 340 360 360 360 360 380 380 380
[39] 400 400 400 400 400 440 460 480 480 480 480 500#save the speed column as an object
car_speed <- cars$speed- logic and boolean operators such as
==(equal to),!=(not equal to),>=(equal to or greater than) or=<(equal to or less than). The object we saved above is called avector. These are very common and we will come back to vectors later, but here’s how operators work on vectors.TRUEandFALSEare always capital, and cannot be used to name objects.
#logical result in place for each item
car_speed > 20 [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUEcar_speed == 4 | car_speed == 7 #vertical bar = or [1] TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[49] FALSE FALSEcar_speed != 4 [1] FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[13] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[37] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE#retrieve items from a vector with a [] brackets
car_speed[car_speed > 20] [1] 22 23 24 24 24 24 25car_speed[car_speed == 4][1] 4 4- quotation marks in pairs are common: both
'and"are the same, use what you prefer. The backward tick ` has special uses and is not the same as single or double apostrophes. The name of an object stored in the Environment is used without quotation marks.
Importing data from Excel
This is likely to be the most common thing we need to do before analysing data. This way we will also create our first object in R.
First, we will copy and save the two tables on the Data tables page, and then import those tables into RStudio.
RStudio makes this easy through menu options, which only works if the package readxl is installed. In RStudio use the File > Import dataset > From Excel to import a sheet from an Excel file.
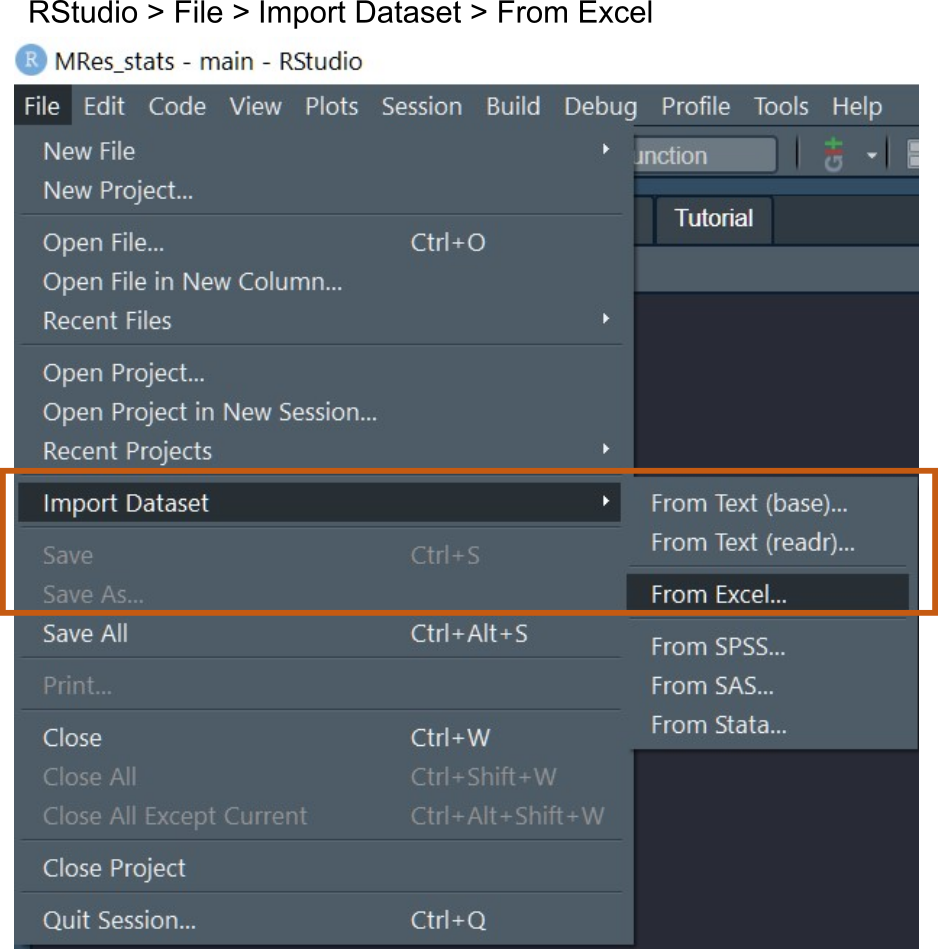
For this session, we will import a wide and long data table, which are saved in Sheet 1 and Sheet 2 of an Excel file called “Weights.xlsx”.
Excel file
You can download these “Mouse Weights” data as an Excel file from here.
Once you have the Excel file, import it into RStudio as follows. If all goes well, the code will appear on the console.
This is the code if data are imported ‘programmatically’ instead of the menu options in RStudio. Note the <- operator that assigns the output of the read_excel function with the argument path assigned using = to the name of the file we want to import given within double quote marks. We use the sheet argument to indicate which sheet to import (if we don’t, the default “Sheet 1” will be imported).
#start the package readxl
library(readxl)
#use the read_excel function
#name the imported table in Sheet1 as Weights
Weights <- read_excel(path = "Weights.xlsx")
#name the imported table in Sheet2 as Weights_long
Weights_long <- read_excel("Weights.xlsx",
sheet = "Sheet2")These tables should appear as objects in the Environment pane of RStudio.

data.frame and its properties
Tables, also called data frames, are a very common type of object in R. This is an R object of class data.frame (examples of other classes are matrix and array). A tibble is a newer type of data.frame with some more features.
A vector is the most basic structure type in R, and it makes up other types of objects like data frames and lists.
str, dim, head and $
Common ways of checking the integrity and details of a data table are through these commands.
#structure of the Weights table
str(Weights) tibble [10 × 3] (S3: tbl_df/tbl/data.frame)
$ Mouse: chr [1:10] "A" "B" "C" "D" ...
$ chow : num [1:10] 29.9 27.3 24 24.8 25.2 ...
$ fat : num [1:10] 28.3 29 29 24.6 34.6 ...#structure of the Weights_long table
str(Weights_long) tibble [20 × 3] (S3: tbl_df/tbl/data.frame)
$ Diet : chr [1:20] "chow" "chow" "chow" "chow" ...
$ Bodyweight: num [1:20] 29.9 27.3 24 24.8 25.2 ...
$ Mouse : chr [1:20] "A" "B" "C" "D" ...#dimensions of the table
dim(Weights)[1] 10 3#first 6 rows of the table
head(Weights)# A tibble: 6 × 3
Mouse chow fat
<chr> <dbl> <dbl>
1 A 29.9 28.3
2 B 27.3 29.0
3 C 24.0 29.0
4 D 24.8 24.6
5 E 25.2 34.6
6 F 20.4 28.1# the $ operator to select values in a column
Weights$Mouse [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J"Making a new column in an existing table is quite easy with the <- assign operator. Let’s say we want to create a column that contains the ratio of the body mass from the chow and fat columns.
#create a new column called ratio
Weights$ratio <- Weights$chow/Weights$fat
head(Weights)# A tibble: 6 × 4
Mouse chow fat ratio
<chr> <dbl> <dbl> <dbl>
1 A 29.9 28.3 1.06
2 B 27.3 29.0 0.942
3 C 24.0 29.0 0.828
4 D 24.8 24.6 1.01
5 E 25.2 34.6 0.729
6 F 20.4 28.1 0.725Columns as factors for t-tests and ANOVAs
Student’s t test (when comparing exactly 2 groups) and ANOVAs (analysis of variance; used when there are more than 2 groups being compared) require groups as discreet or categorical variables (e.g., Untreated & Treated, WT vs KO, Boys vs Girls).
In R, such categorical variables should be encoded as special data format called factor. In statistics as well, we call categorical variables as “factors”, which can have different “levels”. For example, the factor Treatment can have levels such as “Untreated” and “Treated” or more. The factor Genotype can have levels “WT”, “KO”, “Complement”.
Therefore, columns containing categorical data should be explicitly converted into factor type using the as.factor function.
In the long data table (Weights_long), we have one factor (Diet) with two levels (“chow” and “fat”), and corresponding observations of Bodyweight fall within these two levels. “Mouse” is also a factor with 10 levels (A-J).
#run as.factor and assign back to same column
Weights_long$Diet <- as.factor(Weights_long$Diet)
Weights_long$Mouse <- as.factor(Weights_long$Mouse)
#check the structure
str(Weights_long) #compare with previous result abovetibble [20 × 3] (S3: tbl_df/tbl/data.frame)
$ Diet : Factor w/ 2 levels "chow","fat": 1 1 1 1 1 1 1 1 1 1 ...
$ Bodyweight: num [1:20] 29.9 27.3 24 24.8 25.2 ...
$ Mouse : Factor w/ 10 levels "A","B","C","D",..: 1 2 3 4 5 6 7 8 9 10 ...#check levels within a factor
levels(Weights_long$Diet) #"chow" and "fat"[1] "chow" "fat" #number of levels
nlevels(Weights_long$Diet)[1] 2By default, R will order levels alphabetically. This order will also be followed for graphs (i.e., “KO” will be plotted before “WT”), but can be changed using arguments to the factor function.
#change order of levels to "fat" & "chow"
Weights_long$Diet <- factor(Weights_long$Diet, #select column Diet
levels = c("fat", "chow")) #vector in desired order
#check structure
str(Weights_long)tibble [20 × 3] (S3: tbl_df/tbl/data.frame)
$ Diet : Factor w/ 2 levels "fat","chow": 2 2 2 2 2 2 2 2 2 2 ...
$ Bodyweight: num [1:20] 29.9 27.3 24 24.8 25.2 ...
$ Mouse : Factor w/ 10 levels "A","B","C","D",..: 1 2 3 4 5 6 7 8 9 10 ...factor and as.factor are somewhat “advanced” functions while beginning R, but they are required to perform ANOVAs correctly!
It is advisable to get into the habit of converting columns with categorical variables into factors immediately after importing the data frame into R. This is important for calculating ANOVAs correctly.
Read more about factors in the R for Data Science book and on this link to software carpentry.
Functions and arguments
Operations in R are carried out by calling various “functions”. Help on a function can be obtained by typing a ? and name of the function in the console.
Most functions will have arguments, or user-changeable settings that the function accepts. Several of them will have ‘default’ settings that will work in most scenarios. For example, the head function above by default shows 6 rows. This can be changed as necessary by using the argument n (for number of rows). n must always be an integer, otherwise there will be an error!
head(Weights, n = 2)# A tibble: 2 × 4
Mouse chow fat ratio
<chr> <dbl> <dbl> <dbl>
1 A 29.9 28.3 1.06
2 B 27.3 29.0 0.942Objects in R
We encountered tables (data.frame) above. Examples below include vectors and lists.
Vectors and lists
Other very common types of objects in R are a vector and list. A vector is a collection of items, but they have to be of the same type e.g., characters, numbers, logical operators (TRUE or FALSE), factors.
A list can be a collection of vectors or lists, and can therefore contain different types of items.
#a single character
variable1 <- "A"
#combine items into one variable vector with `c()`
#a character vector
vector1 <- c("A", "A", "B", "C", "C")
#check its type
class(vector1)[1] "character"#data.frame columns are a type of vectors
vector2 <- Weights$chow
#vector2 is numeric
class(vector2)[1] "numeric"#vector manipulation
vector2*2 [1] 59.86 54.62 48.06 49.66 50.36 40.72 45.48 47.06 52.20 64.04#coercing a vector type
vector3 <- as.character(vector2)
class(vector3)[1] "character"#create a list with list() function
list1 <- list(vector1, vector2)
list1[[1]]
[1] "A" "A" "B" "C" "C"
[[2]]
[1] 29.93 27.31 24.03 24.83 25.18 20.36 22.74 23.53 26.10 32.02#see its class attribute
class(list1)[1] "list"#numeric vector converted into factor
vector3 <- as.factor(vector2)
str(vector3) Factor w/ 10 levels "20.36","22.74",..: 9 8 4 5 6 1 2 3 7 10#numeric operation will now fail
vector3*2Warning in Ops.factor(vector3, 2): '*' not meaningful for factors [1] NA NA NA NA NA NA NA NA NA NARetrieving items from vectors
#item 2 from vector1
vector1[2][1] "A"#subsetting a vector
vector1[vector1 != "C"][1] "A" "A" "B"#subsetting for items > 29
vector2[vector2 > 29][1] 29.93 32.02Subsetting vectors in-place
#logical operations on character vector
vector1 == "A" [1] TRUE TRUE FALSE FALSE FALSE#numeric logical operations on vector2
#items that match condition will return TRUE
vector2 > 29 [1] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUESubsetting data frames
For data frames, a comma separates subsetting of rows and columns in the data.frame[row, column] indexing format.
To select rows based on a condition, first select the name of the column to set the condition with $. If this part is left blank, all rows are selected.
#rows where chow > 25
Weights[Weights$chow > 25,]# A tibble: 5 × 4
Mouse chow fat ratio
<chr> <dbl> <dbl> <dbl>
1 A 29.9 28.3 1.06
2 B 27.3 29.0 0.942
3 E 25.2 34.6 0.729
4 I 26.1 27.7 0.944
5 J 32.0 35.0 0.914#rows where chow > 25 and column fat in output
Weights[Weights$chow > 25, "fat"]# A tibble: 5 × 1
fat
<dbl>
1 28.3
2 29.0
3 34.6
4 27.7
5 35.0#with a vector of column names
Weights[Weights$chow > 25, c("Mouse", "fat")]# A tibble: 5 × 2
Mouse fat
<chr> <dbl>
1 A 28.3
2 B 29.0
3 E 34.6
4 I 27.7
5 J 35.0#all rows in column Mouse
Weights[, "Mouse"]# A tibble: 10 × 1
Mouse
<chr>
1 A
2 B
3 C
4 D
5 E
6 F
7 G
8 H
9 I
10 J Subsetting and retrieving items from lists
This is similar to handling vectors, but you may need to use [[ double brackets to first select the item in the list.
#retrieving item 3 from list 1 in list1
#note the double [ (`[[`)
list1[[1]][3][1] "B"#retrieve item and operate on it
list1[[2]][3]*2[1] 48.06#subsetting a list
#[[ ]]to get the sub-vector, then subsetting with []
list1[[1]][list1[[1]] != "A"][1] "B" "C" "C"#logical operations on lists
list1[[1]] == "A"[1] TRUE TRUE FALSE FALSE FALSEPlotting a graph
Using the package grafify
For simplicity we will use grafify instead of ggplot2. The grafify package provides a set of functions that calls ggplot2 to plot graphs with fewer lines of code. This is handy for repeatedly plotting graphs while exploring data.
grafify
There will be an error if you have not installed grafify and/or its dependencies, first install the packages, then follow the code below.
#start the grafify package
library(grafify)Loading required package: ggplot2Warning in check_dep_version(): ABI version mismatch:
lme4 was built with Matrix ABI version 1
Current Matrix ABI version is 2
Please re-install lme4 from source or restore original 'Matrix' package#box and whisker plot
plot_scatterbox(data = Weights_long,
xcol = Diet,
ycol = Bodyweight)
#lots of further optimisations are possible
#see https://grafify-vignettes.netlify.appUsing the package ggplot2
This is a simple code to produce a box and whiskers plot; lots and lots of tweaks are possible and almost every little thing can be changed from ‘default’.
ggplot(data = Weights_long,
aes(x = Diet, y = Bodyweight))+
geom_boxplot(aes(fill = Diet))+
geom_jitter()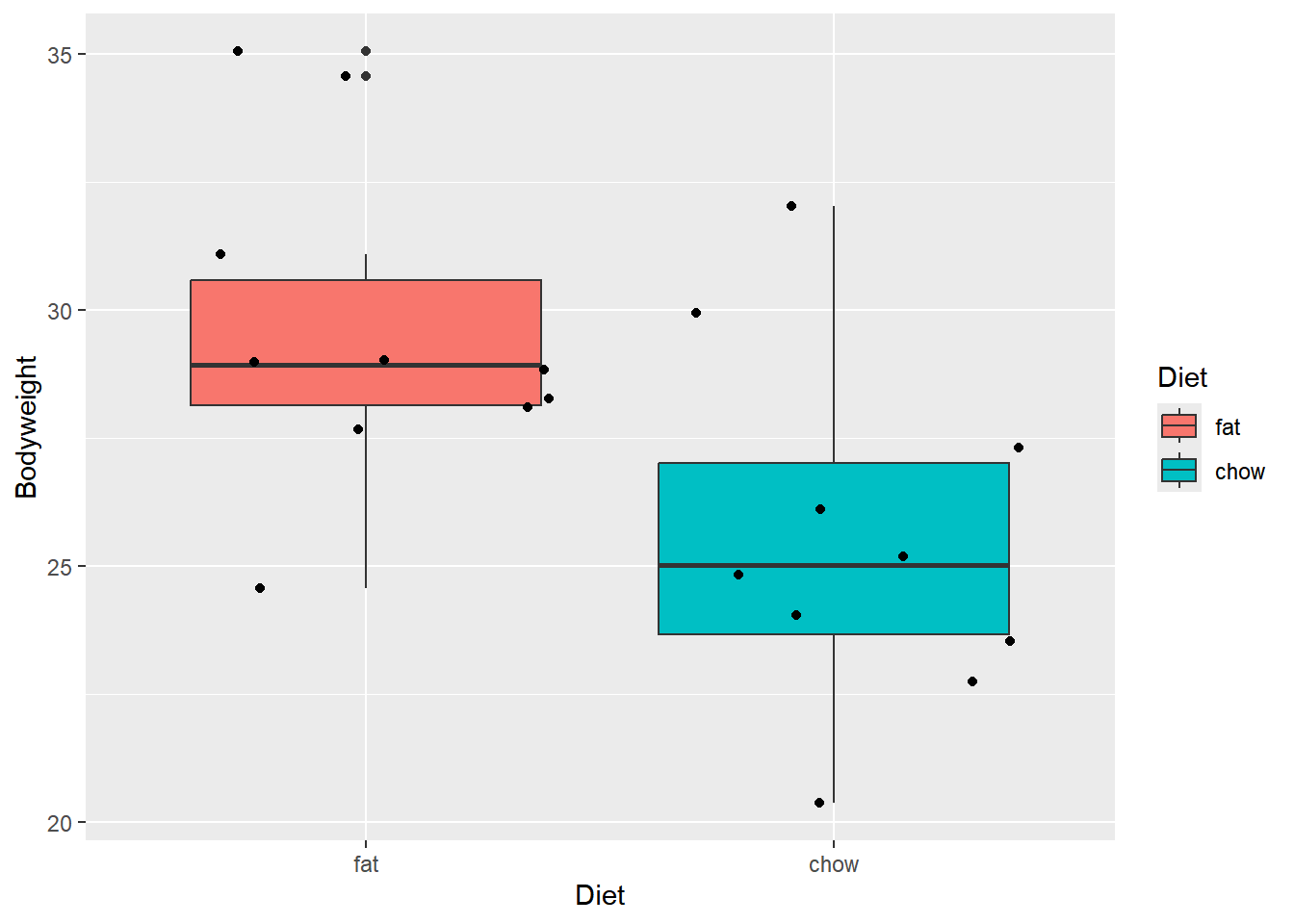
Student’s t test
Formula input with long table
Let’s perform a simple t test in R. The standard ‘formula’ notation is Y ~ X for data in long format. This is read as Y is predicted by X. Y is the dependent variable (Bodyweight in this case), which is predicted by the independent variable, or categorical factor, X (Diet in this case).
The symbol ~ (tilde) has special meaning in R and is used in formulas as above to separate the dependent and independent variables.
t.test(Bodyweight ~ Diet,
data = Weights_long)
Welch Two Sample t-test
data: Bodyweight by Diet
t = 2.7058, df = 17.892, p-value = 0.01453
alternative hypothesis: true difference in means between group fat and group chow is not equal to 0
95 percent confidence interval:
0.8941633 7.1178367
sample estimates:
mean in group fat mean in group chow
29.609 25.603 Column names with wide table
If data are in wide format, the same t.test function can be used with first two arguments as the two columns to be tested. Note that the results are the same.
t.test(Weights$chow, Weights$fat)
Welch Two Sample t-test
data: Weights$chow and Weights$fat
t = -2.7058, df = 17.892, p-value = 0.01453
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-7.1178367 -0.8941633
sample estimates:
mean of x mean of y
25.603 29.609 Table summaries
There are many ways of getting summaries such as mean and SD for variables in a table. The most common way is to use the tidyverse way. In addition, grafify as a simple function that provides summaries based on groups.
Tidyverse method with dplyr
#start the dplyr package
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionWeights_long %>% #%>% is called the pipe
group_by(Diet) %>% #variable to group by
summarise(Mean = mean(Bodyweight), #mean
SD = sd(Bodyweight), #SD
Median = median(Bodyweight), #Median
Count = length(Bodyweight)) #Count# A tibble: 2 × 5
Diet Mean SD Median Count
<fct> <dbl> <dbl> <dbl> <int>
1 fat 29.6 3.18 28.9 10
2 chow 25.6 3.44 25.0 10grafify method
There is simple function called table_summary in grafify to do this.
table_summary(data = Weights_long, #table name
Ycol = "Bodyweight", #numeric variable
ByGroup = "Diet") #grouping variable(s) Diet Bodyweight.Mean Bodyweight.Median Bodyweight.SD Bodyweight.Count
1 fat 29.609 28.915 3.179509 10
2 chow 25.603 25.005 3.436659 10Today’s code for Rmarkdown
Copy all following lines of code inside an R code chunks in the .rmd file. You can add plain or rich text as you like outside the code to describe the code!
#start the package readxl
library(readxl)
#use the read_excel function
#name the imported table in Sheet1 as Weights
Weights <- read_excel("Weights.xlsx")
#name the imported table in Sheet2 as Weights_long
Weights_long <- read_excel("Weights.xlsx",
sheet = "Sheet2")
#start the grafify package
library(grafify)
#box and whisker plot
plot_scatterbox(data = Weights_long,
xcol = Diet,
ycol = Bodyweight)
#perform a t.test
t.test(Bodyweight ~ Diet,
data = Weights_long)If all goes OK, you should see the ‘output’ and code in the new R Markdown file in the Viewer pane. The R Markdown and HTML files are your record of the code used for the analyses and its output (along with the raw data in the Excel file).
R Markdown files will only work if all objects required for the code are created and/or imported within that file. It will not “look for” objects in your Environment - objects have to be created by the code within the R Markdown. This is by design. It is a good feature because if you share your R Markdown file with colleagues, it will still work!
Similarly, ‘kitting’ an R Markdown file does not create any new objects in your Environment.
In the examples from session 1, the “Weights” data frame needs to be imported and created within the R Markdown document even though you might already have it in your Environment. Without the lines of code that import the data, the rest of the code will not work!
Happy R’ing!